Graphics with the mosaic and lattice packages
2023-11-10
Source:vignettes/web-only/GraphicsWithMosaic.Rmd
GraphicsWithMosaic.RmdThis vignette is simply a suite of plots that exist primarily as part of our quality control for the package. But since the examples might be useful to others as well, we’ve added this as a vignette in the package.
This way of doing this is largely superceded by our
ggformula package which provides a formula interface to
ggplot2. You might also like to see the vignette
that compares using lattice to using
ggformula.
lattice extras
The mosaic package resets the default panel function for
histograms. This changes the default for bin selection and provides some
additional arguments to histogram.
ladd()
ladd() provides a relatively easy way to add additional
things to a lattice graphic.
xyplot( rnorm(100) ~ rnorm(100) )
ladd( grid.text("Here is some text", x=0, y=0, default.units="native") )
ladd( panel.abline( a=0, b=1, col="red", lwd=3, alpha=.4 ) )
ladd( panel.rect(x=-1, y=-1, width=1, height=1, col="gray80", fill="lightsalmon"))
ladd( panel.rect(x=0, y=0, width=2, height=2, col="gray80", fill="lightskyblue"),
under=TRUE)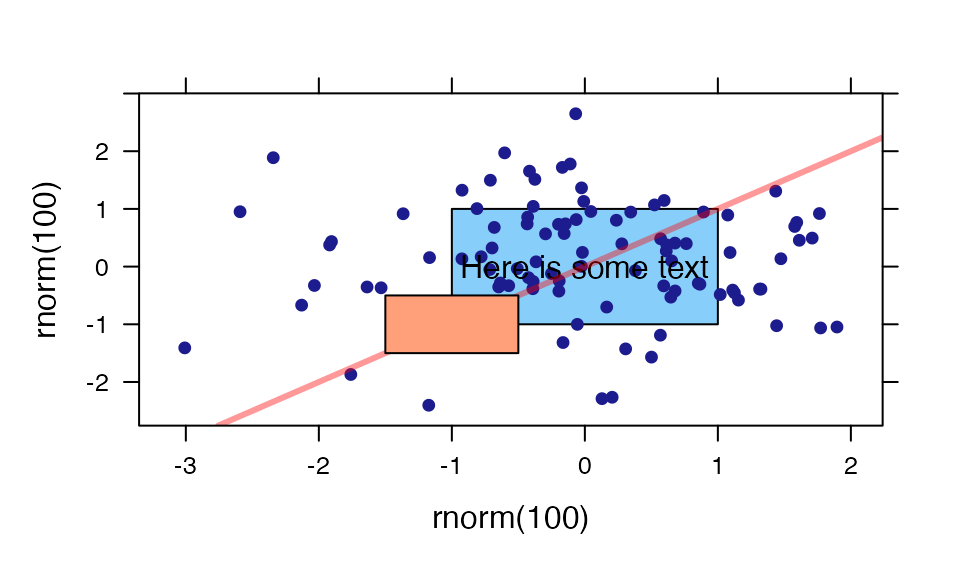
mplot()
In addition to the interactive uses of mplot(), it can
be used in place of plot() in several settings.
require(gridExtra)
mod <- lm(width ~ length * sex, data = KidsFeet)
mplot(mod, which = 1:7, multiplot = TRUE, ncol = 2)## `geom_smooth()` using formula = 'y ~ x'
## `geom_smooth()` using formula = 'y ~ x'
## `geom_smooth()` using formula = 'y ~ x'
## `geom_smooth()` using formula = 'y ~ x'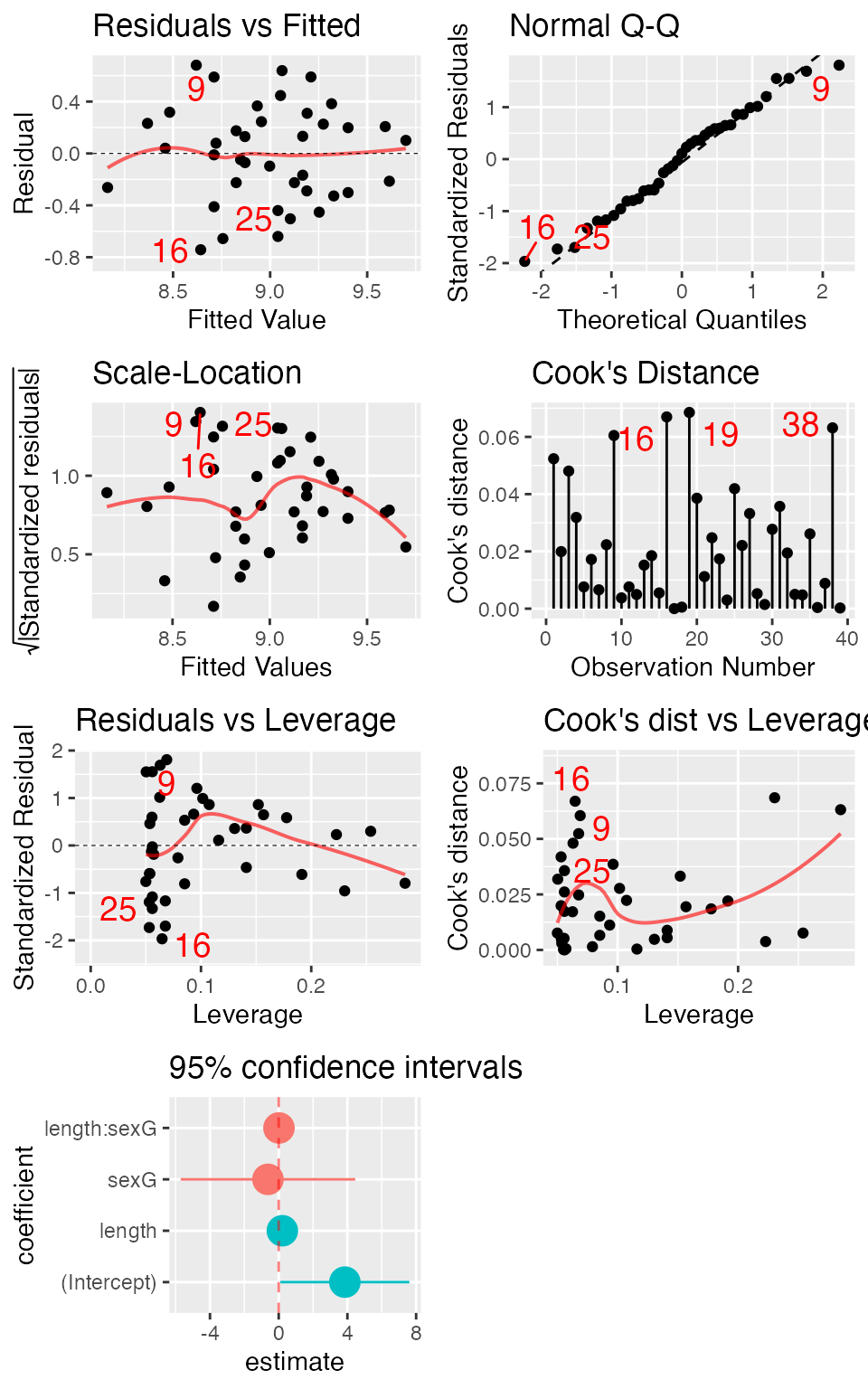
mplot(mod, which=1:7, system="ggplot", ncol=2)## `geom_smooth()` using formula = 'y ~ x'
## `geom_smooth()` using formula = 'y ~ x'
## `geom_smooth()` using formula = 'y ~ x'
## `geom_smooth()` using formula = 'y ~ x'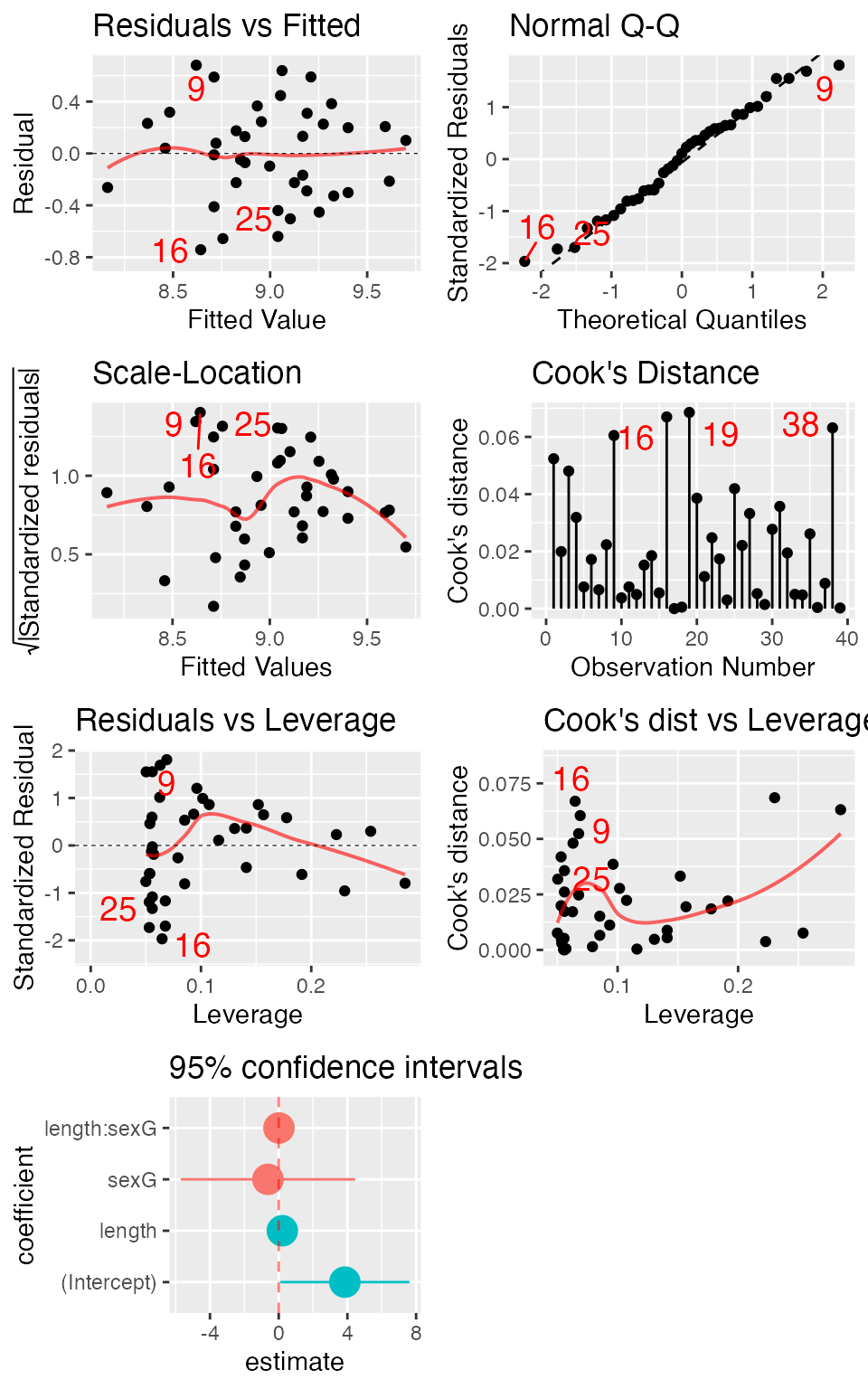
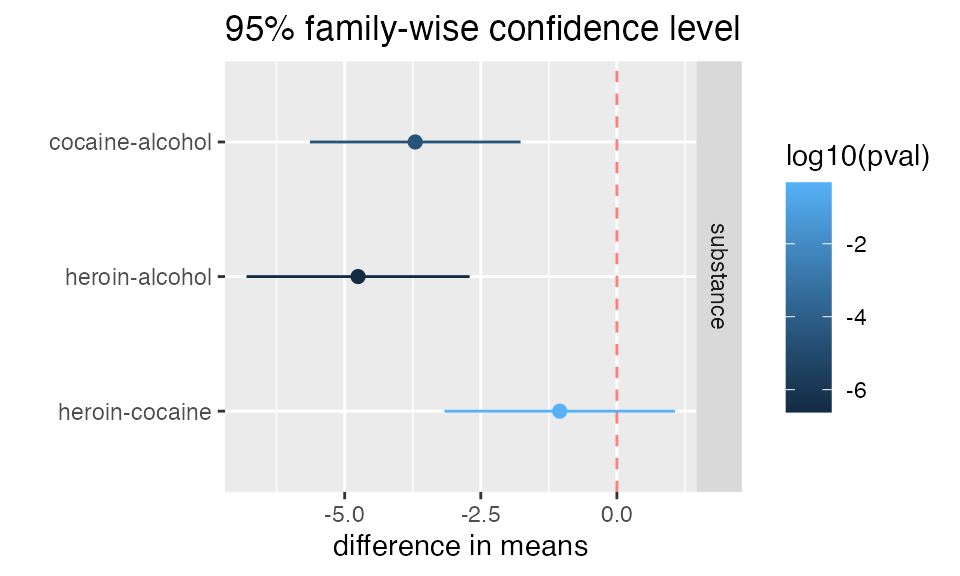
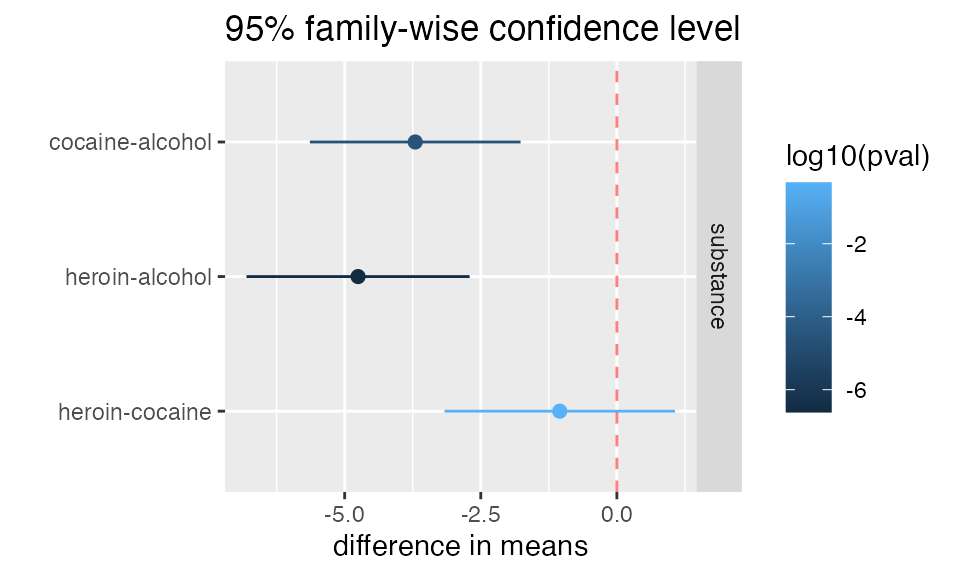
mplot(mod, which=7)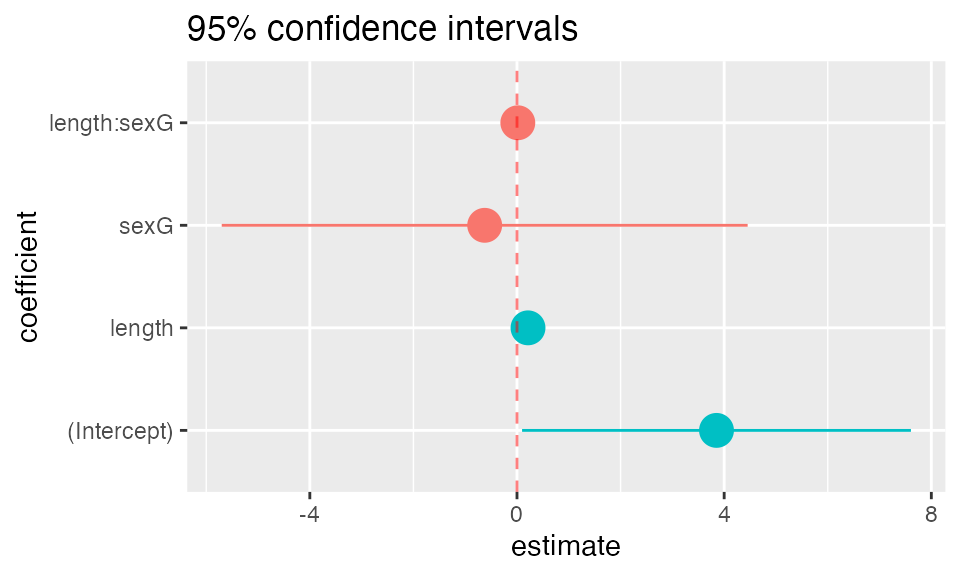
mplot(mod, which=7, rows=-1)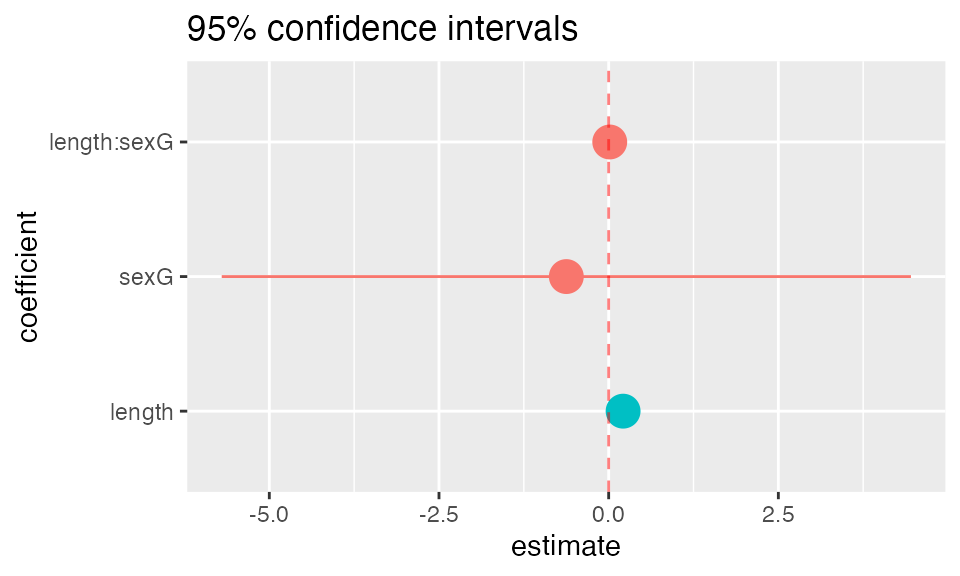
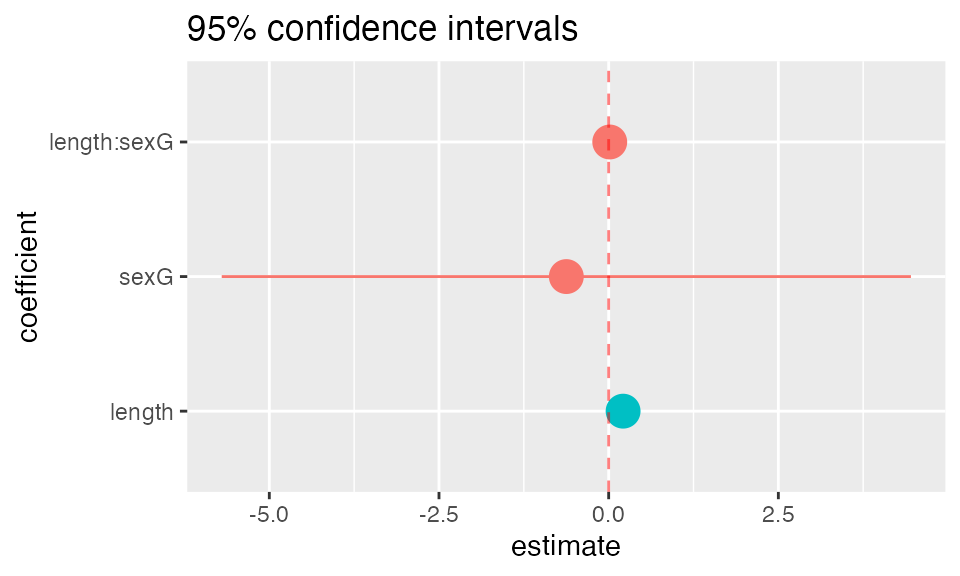
plotFun() and makeFun()
## 1
## 7.041437
L( length=15, sex="G")## 1
## 6.654868
xyplot(width ~ length, groups = sex, data = KidsFeet, auto.key=TRUE)
plotFun( L(length, sex="B") ~ length, add=TRUE, col=1 )## converting numerical color value into a color using lattice settings
plotFun( L(length, sex="G") ~ length, add=TRUE, col=2 )## converting numerical color value into a color using lattice settings
## converting numerical color value into a color using lattice settings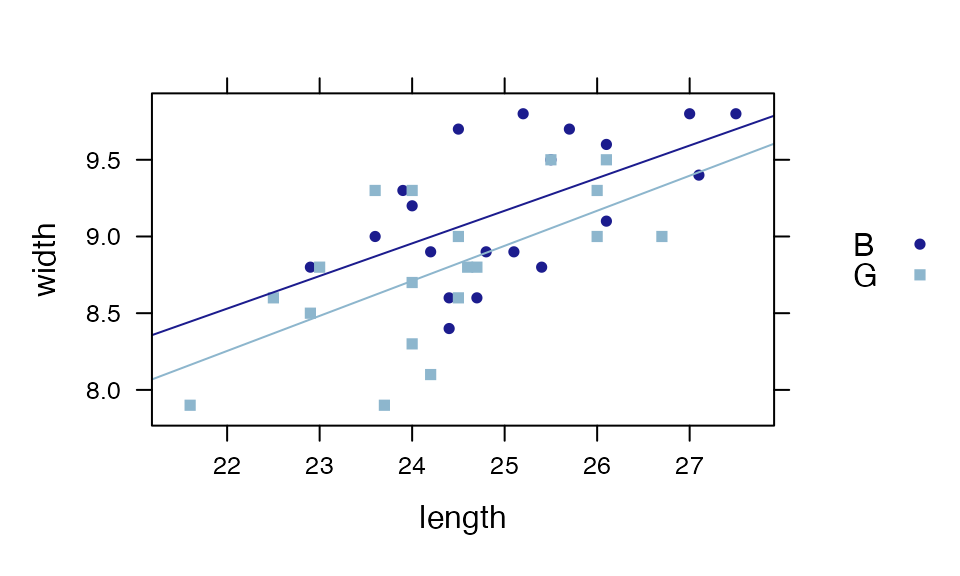
For logistic regression, makeFun() handles the
conversion back to probabilities by default.
mod <- glm( SmokeNow =="Yes" ~ Age + Race3, data=NHANES, family=binomial())
SmokerProb <- makeFun(mod)
xyplot( SmokeNow=="Yes" ~ Age, groups=Race3, data=NHANES, alpha=.01, xlim=c(20,90) )
plotFun(SmokerProb(Age, Race3="Black") ~ Age, col="black", add=TRUE)
plotFun(SmokerProb(Age, Race3="White") ~ Age, col="red", add=TRUE)
ladd(grid.text("Black", x=25, y=SmokerProb(25, Race="Black"),hjust = 0, vjust=-0.2,
gp=gpar(col="black"),
default.units="native"))
ladd(grid.text("White", x=25, y=SmokerProb(25, Race="White"),hjust = 0, vjust=-0.2,
gp=gpar(col="red"),
default.units="native"))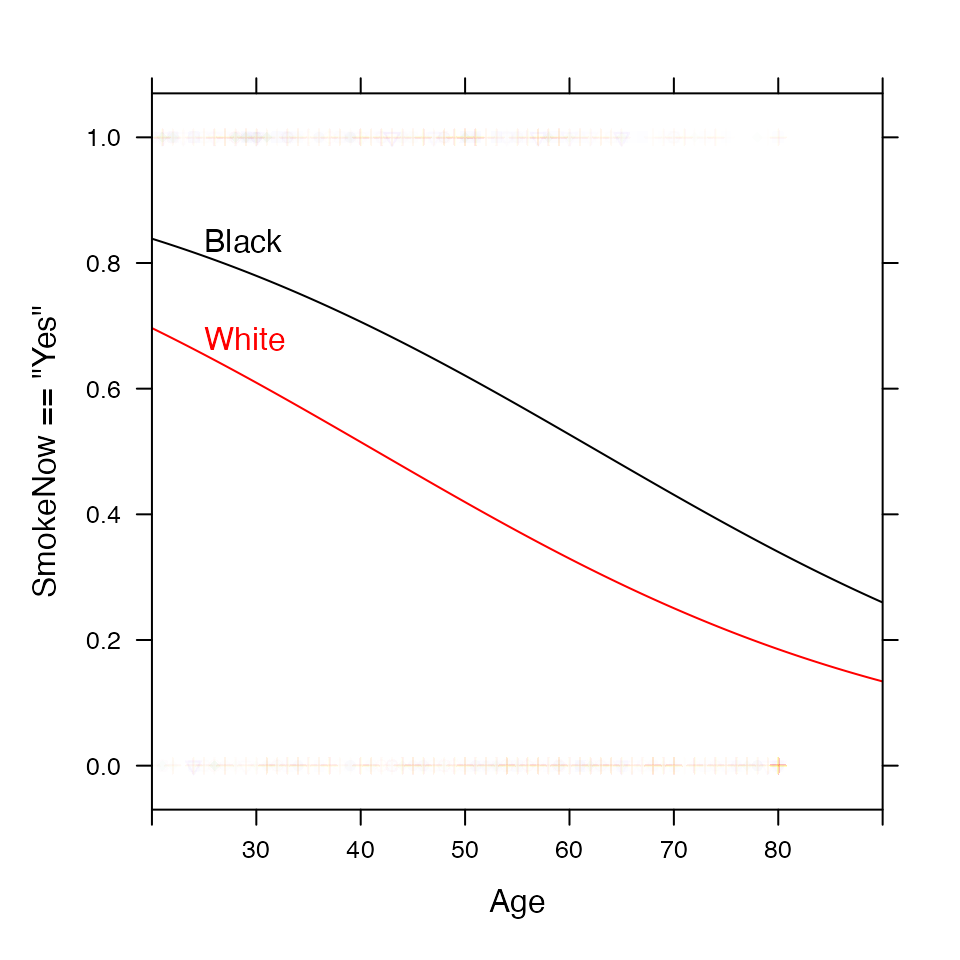
Visualizing distributions
plotDist("chisq", df=3)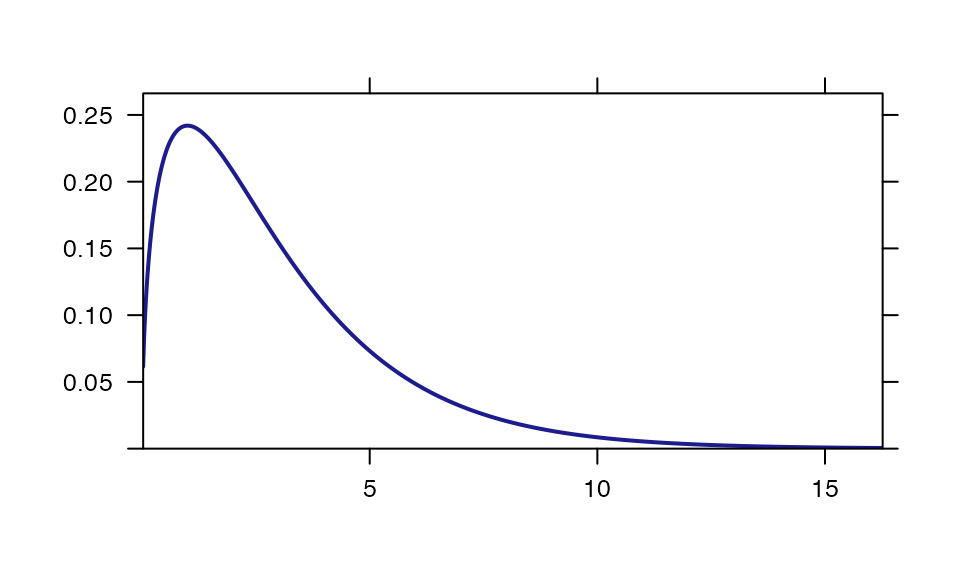
plotDist("chisq", df=3, kind="cdf")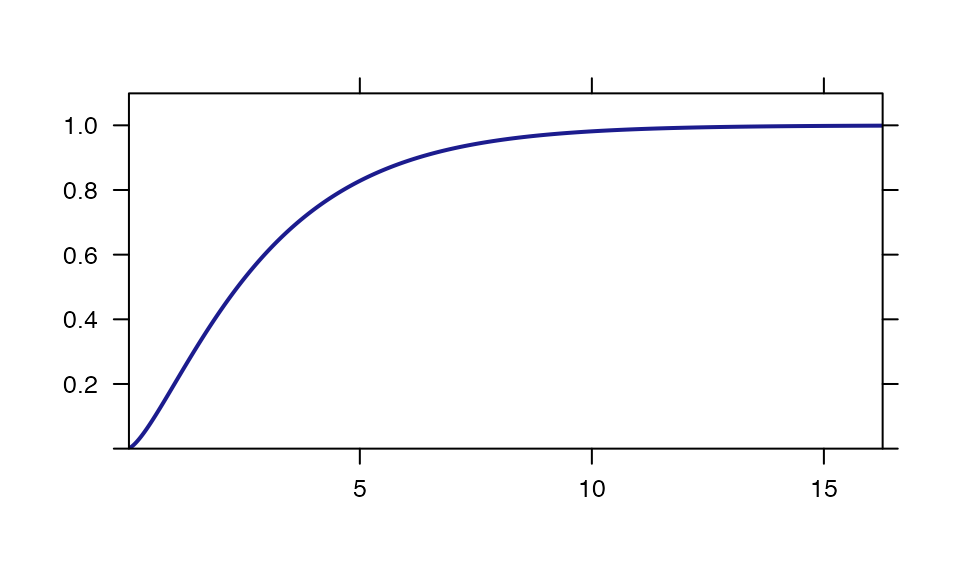
xpnorm(80, mean=100, sd=15)## ## If X ~ N(100, 15), then## P(X <= 80) = P(Z <= -1.333) = 0.09121## P(X > 80) = P(Z > -1.333) = 0.9088## 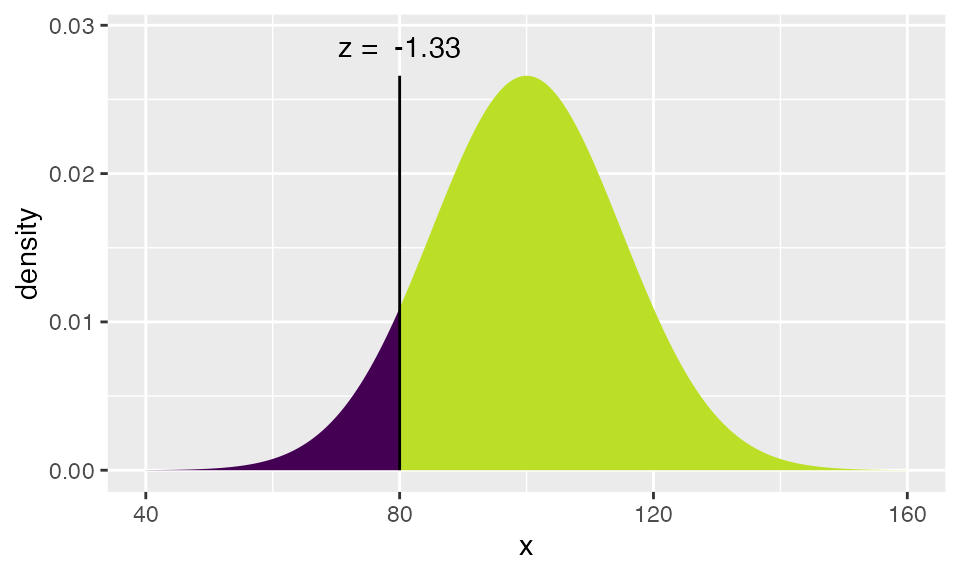
## [1] 0.09121122## ## If X ~ N(100, 15), then## P(X <= 80) = P(Z <= -1.333) = 0.09121 P(X <= 120) = P(Z <= 1.333) = 0.90879## P(X > 80) = P(Z > -1.333) = 0.90879 P(X > 120) = P(Z > 1.333) = 0.09121## 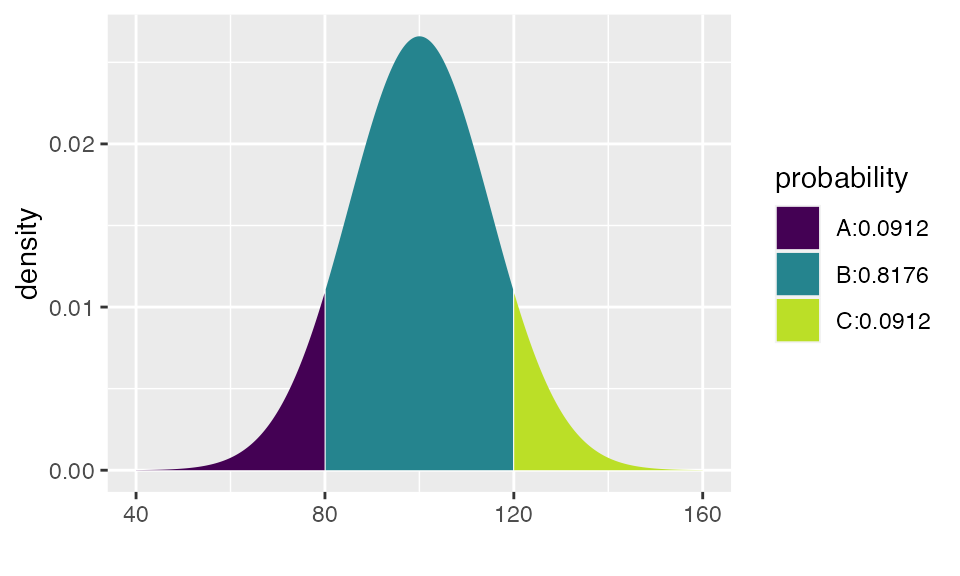
## [1] 0.09121122 0.90878878
pdist("chisq", 4, df=3)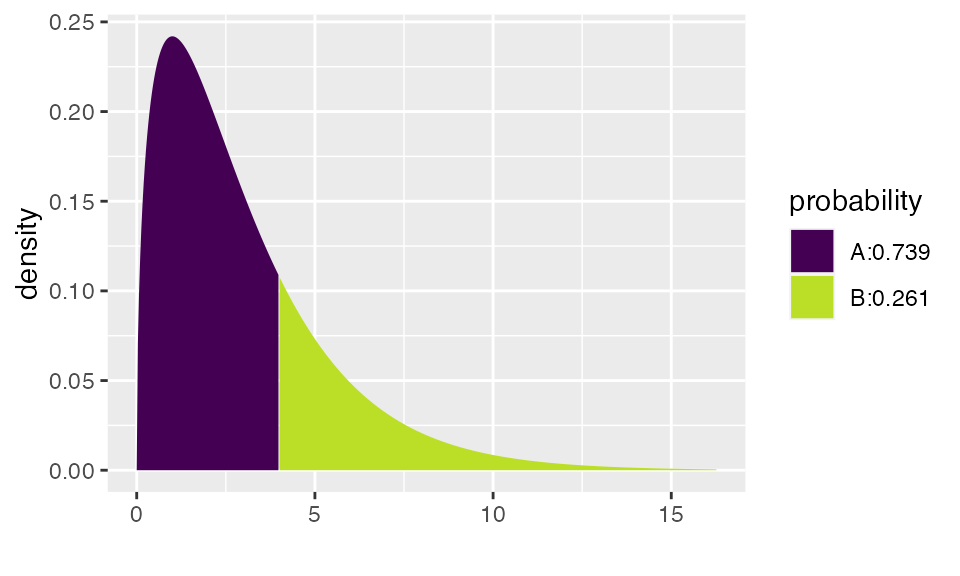
## [1] 0.7385359
pdist("f", 3, df1=5, df2=20)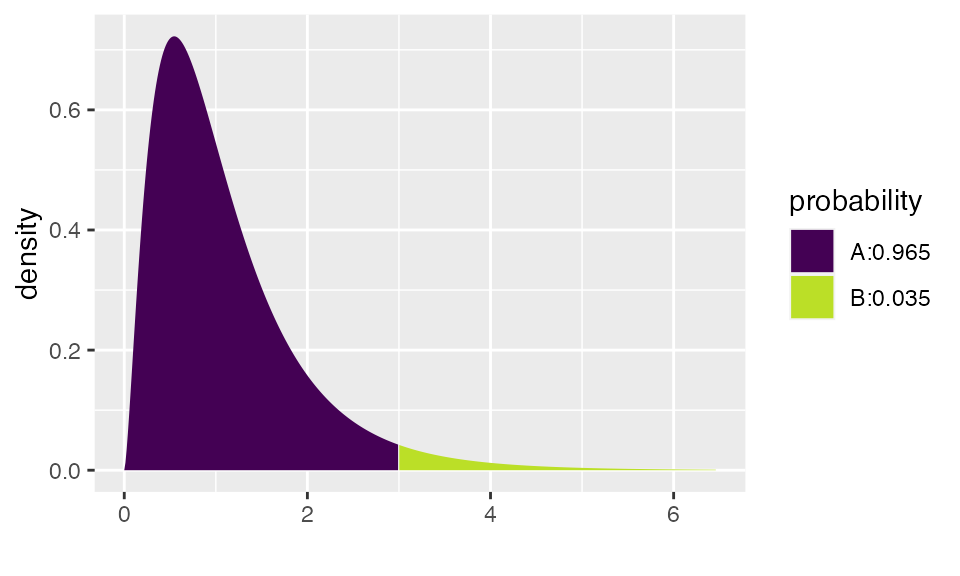
## [1] 0.9647987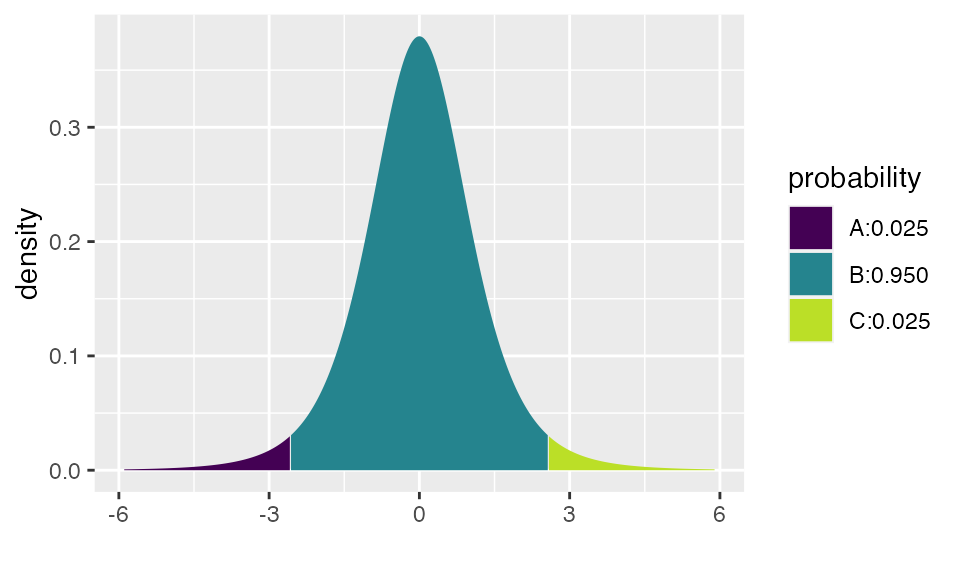
## [1] -2.570582 2.570582
plotDist("norm", col="blue", mean=2, xlim=c(-4,8))
plotDist("norm", mean=5, col="green", kind='histogram', add=TRUE) # add, overtop
plotDist("norm", mean=0, col="red", kind='histogram', under=TRUE) # add, but underneath!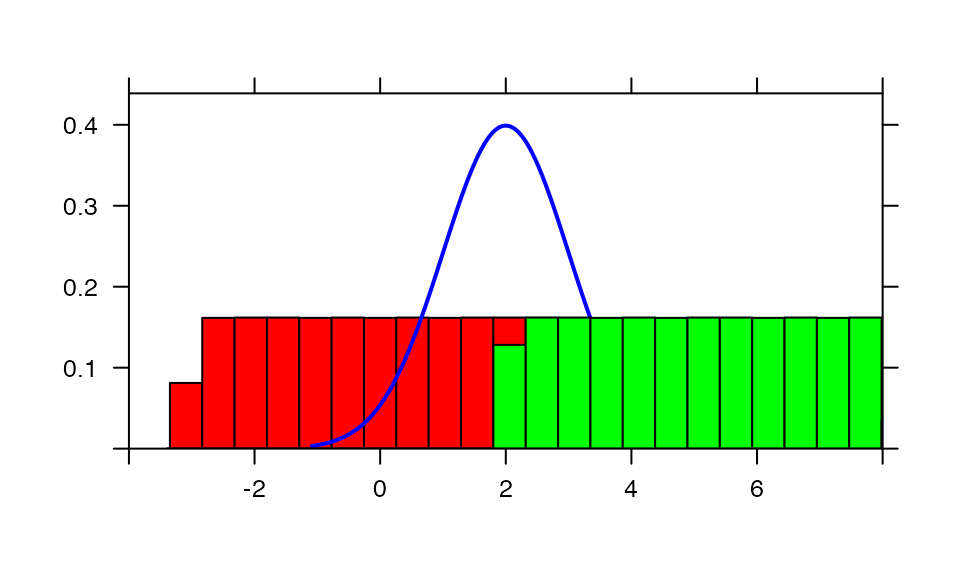
Maps
The mosaic package now provides facilities for producing
choropleth maps. The API is still under developement and may change in
future releases.
## Mapping API still under development and may change in future releases.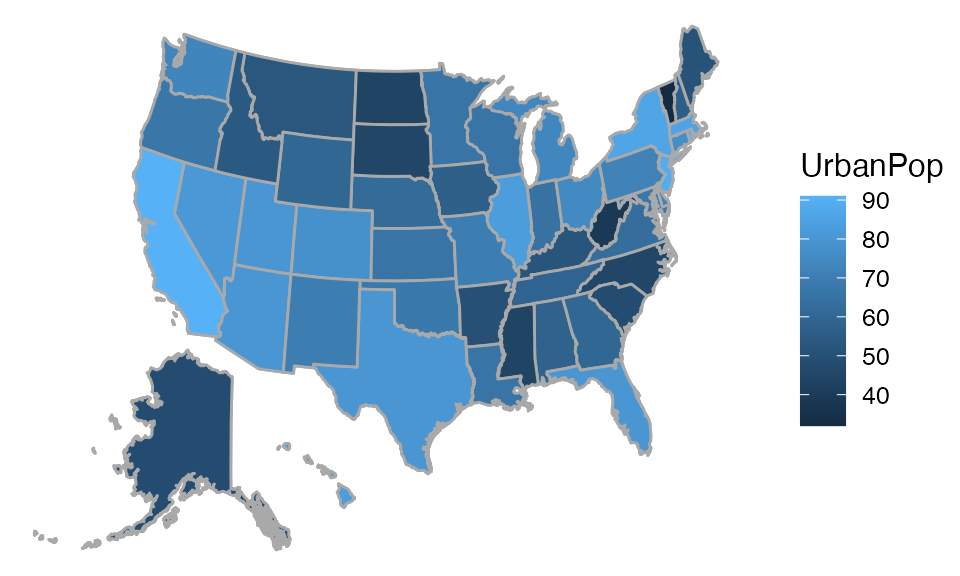
Looks like it is safer to live in the North:
## Mapping API still under development and may change in future releases.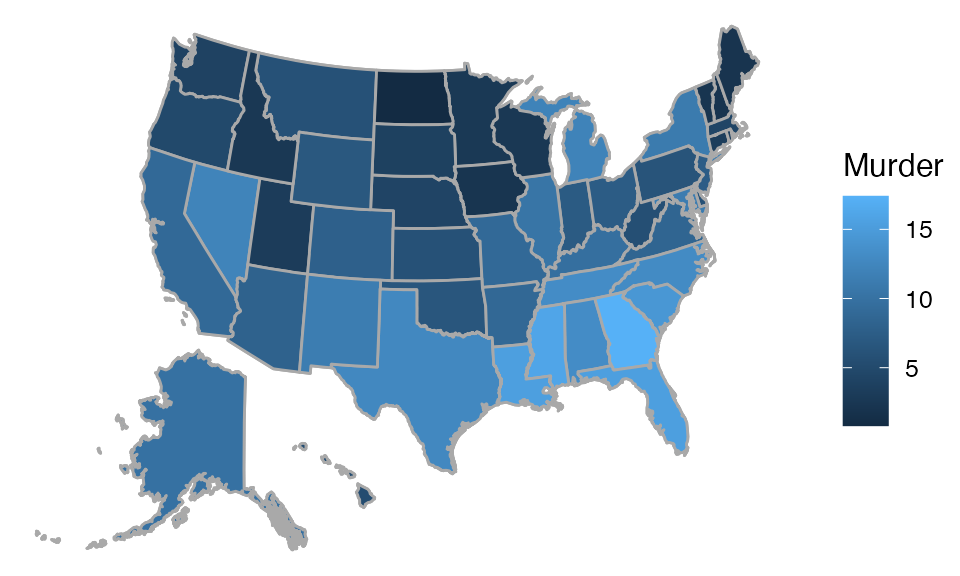
Here is a sillier example
## Mapping API still under development and may change in future releases.## Warning in standardName(x, countryAlternatives, ignore.case = ignore.case, : 99
## items were not translated